MitoEM Dataset: Large-scale 3D Mitochondria Instance Segmentation from EM Images
4Technical University of Munich 5Shanghai Jiao Tong University 6Northeastern University
7Indian Institute of Technology Roorkee 8Ikerbasque, Basque Foundation for Science
[Paper (Updated Results)] [Code] [Dataset]
Abstract
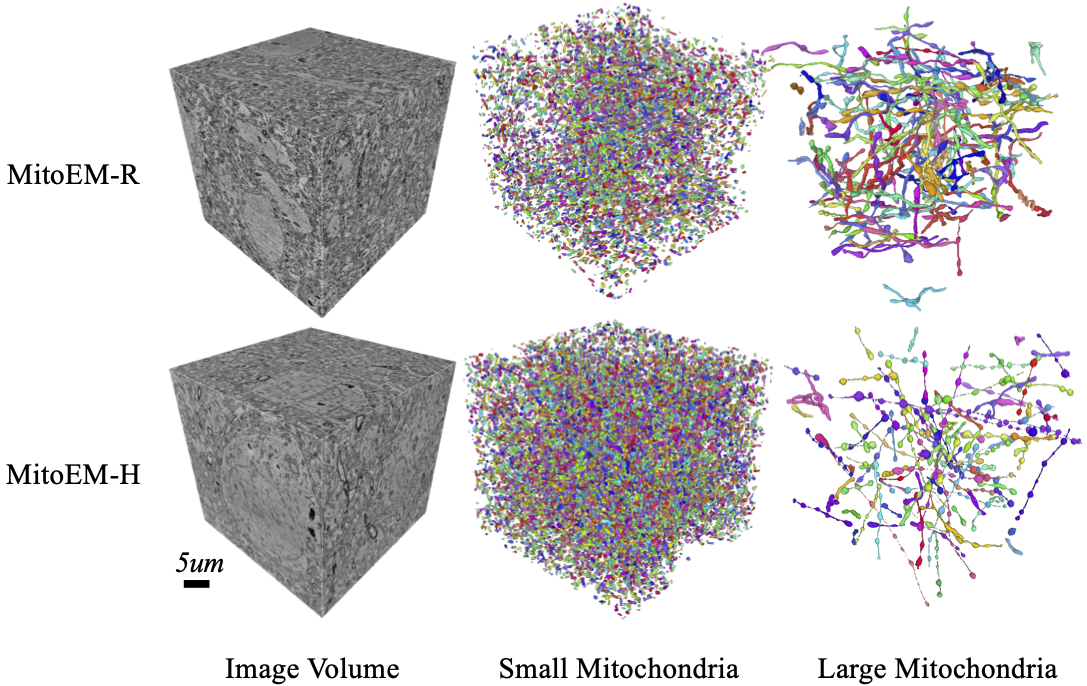
Electron microscopy (EM) allows the identification of intracellular organelles such as mitochondria, providing insights for clinical and scientific studies. However, public mitochondria segmentation datasets only contain hundreds of instances with simple shapes. It is unclear if existing methods achieving human-level accuracy on these small datasets are robust in practice. To this end, we introduce the MitoEM dataset, a 3D mitochondria instance segmentation dataset with two 30x30x30 μm3 volumes from human and rat cortices respectively, 3,600x larger than previous benchmarks. With around 40K instances, we find a great diversity of mitochondria in terms of shape and density. For evaluation, we tailor the implementation of the average precision (AP) metric for 3D data with a 45x speedup. On MitoEM, we find existing instance segmentation methods often fail to correctly segment mitochondria with complex shapes or close contacts with other instances. Thus, our MitoEM dataset poses new challenges to the field.
Dataset
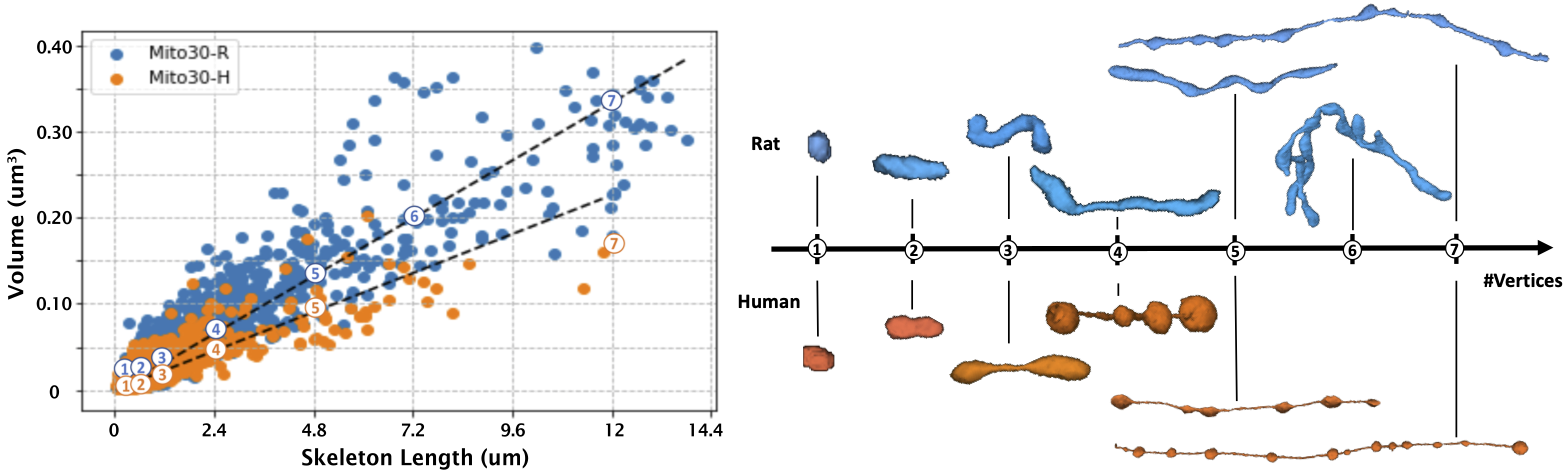
(Left) We plot the length versus volume of mitochondria instances for both volumes, where the length of the mitochondria is approximated by the number of voxels in its 3D skeleton. (Right) There is a strong linear correlation between the volume and length mitochondria in both volumes, which is the average thickness of the instance. While the MitoEM-H has more small instances, the MitoEM-R has more large instances with complex morphologies. We sample mitochondria of different length along the regression line and find instances share similar shapes to MOAS in both volumes.
Updates from the Proceeding Version
- The MitoEM dataset is 1,986x bigger than the Lucchi dataset, instead of the 3,600x claimed in the paper. We mistakenly doubled the factor by plugging in only the size of the Lucchi test data. Lucchi dataset annotates two sub-volumes (each 165*1024*768 in voxels) from the 5*5*5 um image volume, and each of the two MitoEM volumes has 4096*4096*1000 voxels at the 8*8*30 nm resolution. Thus, the factor should be ((4096*4096*1000)*(8*8*30)) / ((165*1024*768)*(5*5*5)) \approx 1986.
- Sec. 2 "Dataset Acquisition": the human data is from the temporal lobe instead of the frontal lobe. The entire human dataset was introduced in [Shapson-Coe et al. 2021]
- After the proceeding, we did another round of annotation cleaning and improved the training of our 3D model. Please cite and compare with the following numbers, if you plan to use this dataset. [Descriptions] [Leaderboard Results]
Method MitoEM-H MitoEM-R Small Med Large All Small Med Large All U2D-B v2 0.106 0.592 0.563 0.566 0.057 0.450 0.300 0.335 U3D-BC v2 0.426 0.838 0.798 0.804 0.311 0.845 0.803 0.816
Citation
@inproceedings{wei2020mitoem,
title={MitoEM Dataset: Large-Scale 3D Mitochondria Instance Segmentation from EM Images},
author={Wei, Donglai and Lin, Zudi and Franco-Barranco, Daniel and Wendt, Nils and Liu, Xingyu and
Yin, Wenjie and Huang, Xin and Gupta, Aarush and Jang, Won-Dong and Wang, Xueying and others},
booktitle={International Conference on Medical Image Computing and Computer-Assisted Intervention},
pages={66--76},
year={2020},
organization={Springer}
}
Acknowledgement
This work has been partially supported by NSF award IIS-1835231 and NIH award 5U54CA225088-03.